Getting started!
RGG can be used in two different ways:
1. RGG as an R package within JGR
The first way to use RGG is by installing it as an R Package in JGR. Here JGR provides the connection to R. JGR is a Java GUI for R.
Installation
1. Download and install R (2.8 or higher is recommended)
2. Install Java (6 or newer)
3. Download JGR 1.5 or higher.
4. Install RGG by executing the following lines within JGR:
install.packages("RGG",repos="http://R-Forge.R-project.org")
Notice that this will work only, for the R version 2.5.x and upper. If you use an older version, please go to the Downloads section and download the appropriate RGG package for your operating system and install it manually.
Then load the RGG package by calling "library(RGG)". This will add a menu "RGG" on the menubar of JGR window (see the screenshot below).
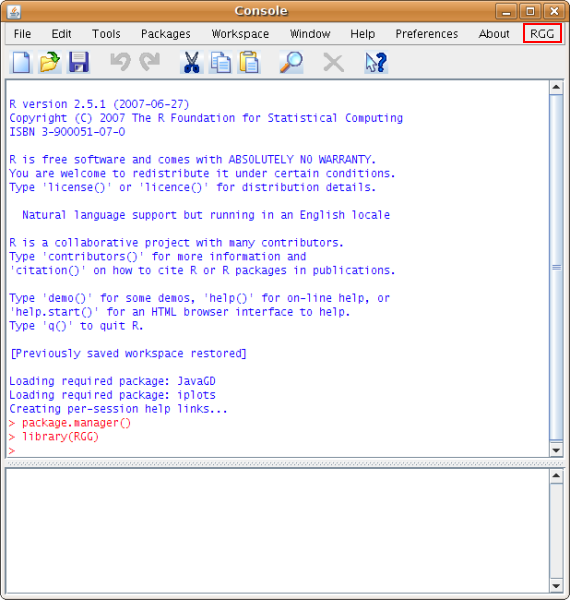
To open an RGG file click on the submenu "Load" of "RGG". Let's load an RGG file, qcreport.rgg.
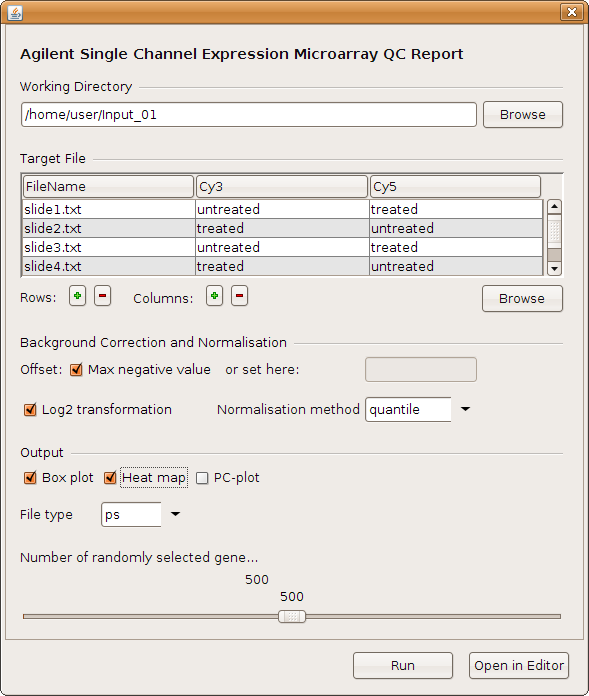
The user interface has two buttons at the bottom: "Run" and "Open in Editor". The "Run" button generates the corresponding R code and sends it to R for execution. "Open in Editor" will open the generated R code in the JGR editor, which uses R specific highlighting.
2. RGGRunner
Installation
1. Download and install R (2.8 or higher is recommended)
2. Install Java (6 or newer)
3.
Install RGG by downloading the executable for your operating system:
Windows: Download the rggrunner-windows.exe
Linux: Download the rggrunner-linux.sh
Mac OS X:
Download the rggrunner-macosx.tgz
Zip distrubition:
rggrunner.zip
The RGGRunner can be
used to open an .rgg file and to execute the generated R script
directly from the RGG.
RGGRunner uses the "Rscript"
command, found in the
/bin directory of R
installation folder, to
execute the
generated R script. If this command is not found in the PATH environment
variable, user will be prompted to enter the installation folder (e.g.
on Windows "C:\Program Files\R\R-2.8.0\bin").
Here is a screenshot of the RGGRunner. An .rgg file
(qcreport.rgg)
is already loaded and executed here.
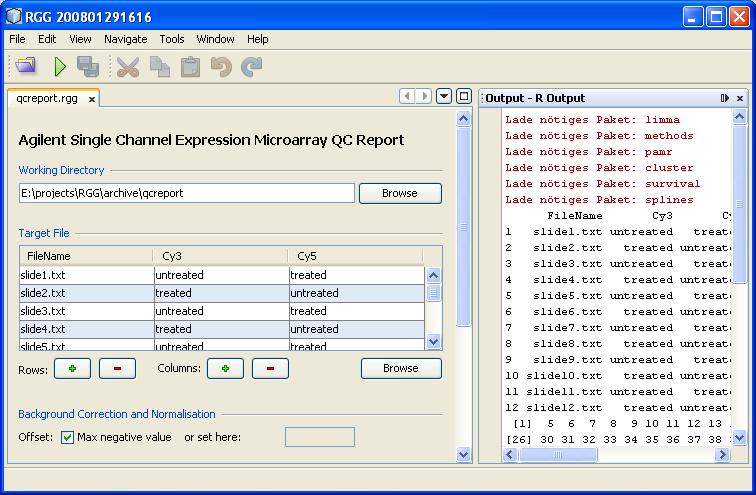
Figure 1: RGGRunner
The main window consists of two parts, left (GUI) and right (output), which can be rearranged by drag and drop. An .rgg file is loaded, either by clicking the "Open RGG" button or using the shortcut "CTRL-O". After setting inputs, parameters etc. on the GUI, the generated R script can be executed by clicking the "Run" button or using the "CTRL-R" shortcut. The output from R is displayed on the "Output" window.

